We need a reality check before we can reach for the stars, says Dr Natalie Thorne at Digital Health Festival 2023.

Since sequencing the first human genome 20 years ago, we’re well and truly in the genomic era. Multiple studies – including several in Victoria – have proven that genomics can deliver more diagnoses and better care, across many health conditions.
The dream is precision medicine: healthcare tailored to your genes and environment, resulting in better health outcomes for you, and massive cost savings to the health system.
But what will it take to shift gears beyond single-use genomics, to multi-use genomics, and then precision healthcare?

Many factors hold genomics back. Data management is one of the biggest.
Around 97% of data generated in healthcare is inaccessible and not reusable. Genomic data is headed in the same direction.
Genomic tests involve moving large, complex datasets between multiple analytics tools. So labs not only have to manually download and upload data from one tool to the next, but they also have to manage the data translation and transformation, and the software testing and updates for each tool.
This means hours of manual work per test. And at the end, the patient’s genomic data ends up in a silo.
So what happens if that data needs to be shared with another lab – say, because the patient has changed hospitals or moved to another state?
Labs first spend hours trying to find the data. Then the blueprints of human beings are ferried around on USB sticks. Not only is that prone to error and unsafe, but it stops genomics from ever being scalable.

So what would it take for labs to be able to flick the stick?
Much like labs now have a LIMS (Laboratory Information Management System) to manage samples and patent information, they will also need a GIMS (Genomic Information Management System) to manage the complex data transfer and storage requirements of genomics.
Melbourne Genomics came to that realisation back in 2013, and we’ve been building our GIMS ever since.
Our software solution, Genomical, is helping labs to scale up genomic testing and protect their patients’ data. But it’s also helping labs to share patient data safely when needed – and that opens the door to new ways of doing genomics.
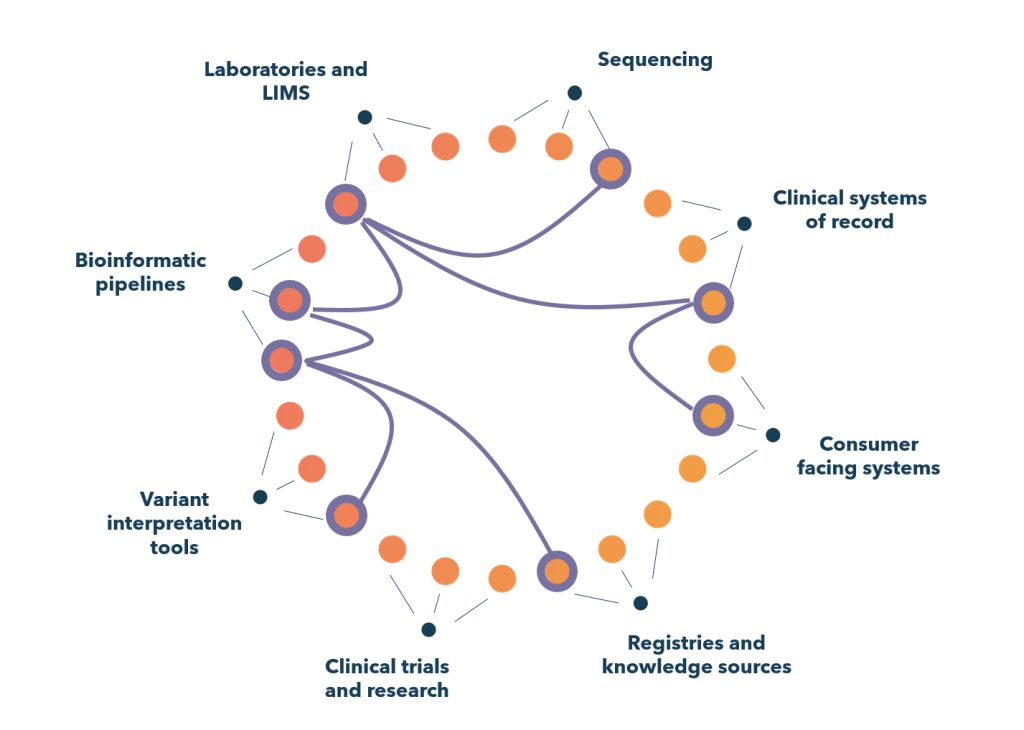
A shared genomic information management system enables connections that are vital for precision healthcare.
Imagine a young girl seeing a paediatrician for a suspected genetic condition. Her parents authorise a genomic test through a consent app. An order goes to the testing laboratory’s LIMS. Then the genomic information management system automates all the data steps required, across different bioinformatics pipelines and variant interpretation tools. It might also enable key results to be anonymously contributed to a global knowledge base.
Now imagine that girl has grown up and needs another test to determine pharmacogenomic risks. The test is done by a different lab – but now she can give the lab permission to access her existing genomic data via the shared system, making analysis far quicker and cheaper.
The ability to connect genomic data into each part of the digital health system is what we’re missing right now. And that’s a complex problem to solve.

Once we use genomic data more effectively, precision medicine become possible.
Labs will be able to better protect our genomic data, and share it when we need them to. New types of tests can be readily rolled out, and new diagnostic products developed to support those tests. Genomic health records can sit comfortably within the health system and guide the care we receive throughout our lives.
A shared ‘GIMS’ ties all of this together. And I’d love to chat further about how to make It happen.
This is an edited version of a presentation delivered by Dr Natalie Thorne at the Digital Health Festival on 6 June 2023.

 News
News
